LCN DNA typing is when the forensic sample has a low amount of template DNA prior to PCR amplification. A low amount of template DNA has been reported as a DNA concentration <100pg (pg = pico grams = one trillionth (10-12) of a gram)1 but in more general terms it is a sample with a DNA concentration less than the manufacture’s recommended DNA template concentration for use with its DNA typing PCR kit. For example, Applied Biosystems recommends 1ng – 2.5ng (1000pg — 2500pg) of template DNA when amplifying DNA samples using the AmpFlSTR® Profiler Plus® kit.2
Has LCN typing been presented in court?
LCN typing results were reported in Australia during the Peter Falconio case (R v Murdoch)3 and in the murder case of the Swedish Foreign Minister, Anna Lindh.4 In the case of the Omagh bombing the Judge questioned the reliability of LCN typing which led to the UK temporarily suspending use of the technique.5
These cases made headlines with the LCN typing terminology but samples have been processed for many other cases where the DNA concentration is below the manufacture’s recommended DNA template concentration for PCR kits. Forensic samples referred to as ‘touch’ or ‘trace’ DNA samples, as well as degraded DNA samples can have a low amount of template DNA prior to PCR amplification. The sensitivity of the PCR conditions for DNA typing PCR kits (e.g. AmpFlSTR® Profiler Plus® kit) can enable detection of alleles (bands) for such samples that are below the manufacture’s recommended DNA template concentration and within the range of LCN DNA. In most instances standard operating protocols for interpretations are used. It is when the standard operating protocols for PCR are modified to increase the detection of LCN DNA template that interpretation of results should be done with caution.6
A reported method to adopt when analyzing a DNA sample of <100pg is to increase the number of PCR cycles from the recommended 28 to 34 cycles.7 DNA results are then analyzed below recommended thresholds where stochastic effects during PCR are observed and standard protocol interpretations are no longer valid.
What are the stochastic effects seen in LCN typing?
A. Differential amplification of heterozygous alleles (bands)
A low amount of template DNA for PCR amplification can cause differential amplification of heterozygous alleles (bands) and result in substantial allele imbalance or allele dropout.
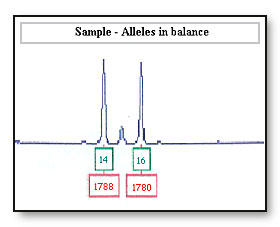
For example: Amplification of the bands 14 and 16 with peak heights of 1788 and 1780 rfus (relative fluorescence units). Validation testing of the PCR kit reports alleles are usually within balance and have a peak height ratio of 70% or higher.8
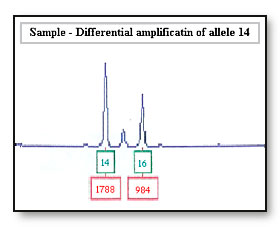
Differential amplification of the 14 allele will be seen if the peak height ratio for the 14 and 16 allele is less than 70%.
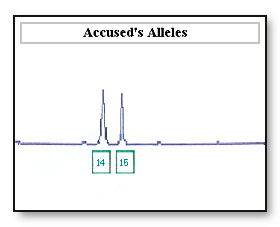
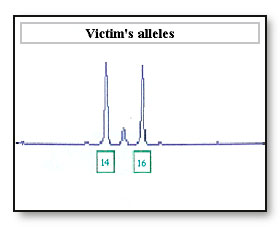
Consideration: In some cases peak height is used to assign bands in a mixed sample. An allele imbalance of the 14 and 16 allele may be wrongly interpreted as a mixed sample, where there are two contributors to the 14 allele and one contributor to the 16 allele
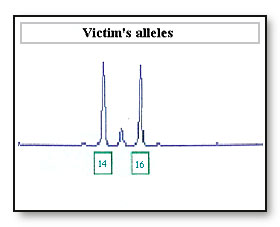
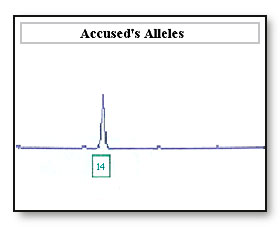
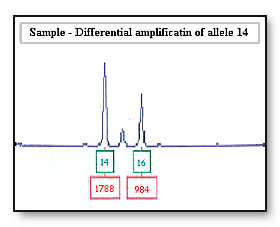
If the 16 allele has dropped out as a result of differential amplification of the 14 allele the sample maybe incorrectly interpreted as a 14, 14 DNA profile at this locus. If the accused is a 14, 14 the accused’s profile will be reported as being not excluded from this locus.
B. Stutter bands
Stutter bands occur in general for all loci. Validation testing of the PCR kit reports stutter bands are observed on the left and below 14% of the true allele.9 Testing using LCN DNA typing have observed stutter bands above 14% in height of the true allele and are sometimes reported at a greater height than the true allele.10
Consideration: If the accused has a band at a stutter band site then this stutter band might be considered a true allele and the accused’s profile will be reported as being not excluded from this locus.
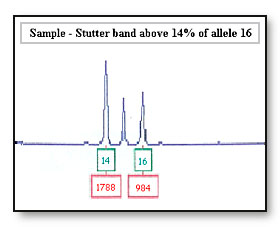
It also might be that the evidence is considered to be a mixed sample that is hard to interpret when in fact it is only one contributor with stutter.
C. Spurious bands
It has been reported that spurious bands have been seen in the negative control, even when testing was performed under strict cleanliness criteria.11
Consideration: Spurious bands (or drop-in alleles) could be considered as a true allele consistent with the accused’s reference sample, increasing the chance of a false positive match.
Questions to ask when LCN DNA typing is presented in your case.
While this evidence may improve with time and laboratory research it is important to be cautious when considering what the DNA profile results mean for your case.
- Identify which DNA typing PCR kit was used by the laboratory.
- Review the PCR method to see the DNA concentration used for the sample, and the number of PCR cycles.
- Do the laboratory procedures include any modifications to the standard operating protocols to increase the sensitivity of detection?
- Identify if the laboratory has performed validation studies for PCR conditions outside the standard operating protocols.
- Is the DNA typing PCR kit subjected to quality control at these conditions?
- Were the alleles interpreted using standard laboratory protocols or laboratory protocols for modified PCR conditions?
- Review negative controls for spurious bands.
- Does the reporting scientist understand the limitations in reporting and interpretation of LCN typing?
- Were the results validated by reproducibility — multiple amplifications of the same sample? Good laboratory practice would include repeating the sample from the same area, and this is a recommendation of Gill in the use of LCN.12 If this is not possible due to a small amount of sample then is it reasonable to rely on such evidence?
Carol Mayne
B.Biomed.Sc PGDipSc,MolGen MSc(Forensic)
Footnotes
- P Gill, et al, ‘An investigation of the rigor of interpretation rules for STRs derived from less than 100pg of DNA’ (2000) 112, 1 Forensic Science International 17-40.
- Applied Biosystems, AmpFlSTR® Profiler Plus® PCR Amplification Kit, User’s Manual (2006) 1-9.
- R v Murdoch (2005) NTSC 76.
- ‘British experts help Swedish manhunt’, BBC News 14 September 2003 <http://news.bbc.co.uk/2/hi/uk_news/england/west_midlands/3107572.stm>.
- The Crown Prosecution Service, ‘Review of the use of Low Copy Number DNA analysis in current cases: CPS statement’ (2008) CPS Press Release <http:cps.gov.uk/news/pressrelease/101_08.html> 14 January 2008.
- B Budowle, et al, ‘Low Copy Number — Consideration and Caution’ (2001) Twelfth International Symposium on Human Identification. Promega Corporation, Madison, Wisconsin, 2001 <http://www.promega.com/geneticidproc/ussymp12proc/default.htm>
- Gill, above n 1, 17.
- Applied Biosystems, above n 2, 9-25
- Budowle, above n 6, 2.
- Gill, above n 1, 17.
- Ibid.
- Ibid.